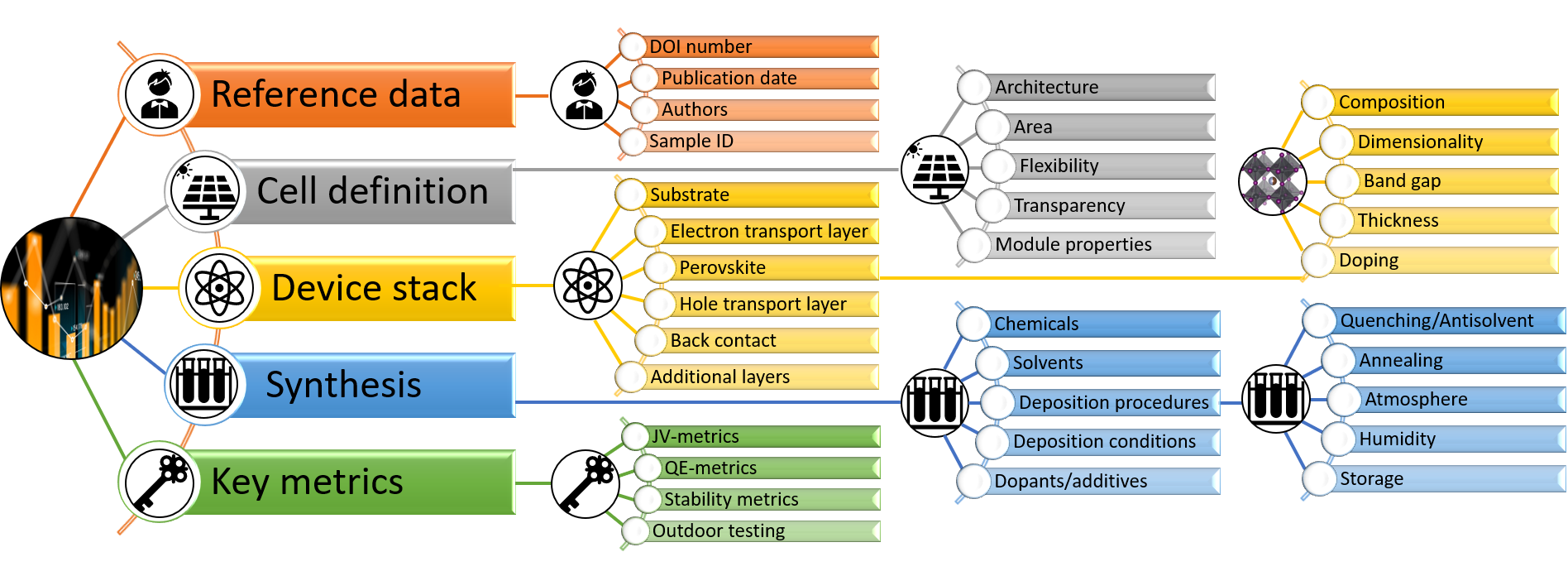
Content of the perovskite database
A detailed description of the categories found in the database can be downloaded below. This include a description of how the data in each category is formatted, when it was implemented into the database, and what we know about accuracy.
The codebase of the project
Most of the code written in the project is in Python, and the interactive graphics is implemented using the Python framework Bokeh.
All code is available at the projects github repository
The code is divided into several sections
- Code for the interactive graphics
- Code for reading in, cleaning, and formatting user data, and for uploading data to the database
- Code for regenerating figures used in papers with the latest data added to the database
- Code for analysing data.
Link to the projects Github repository
If you have written code that utilises the Perovskite Database Project and would like to share it here, please contact the project responsible.
Data extraction protocol
To upload new data to the database, the data must be formatted consistently with respect to the data already in the database. The backend for inserting data to the database consists of an Excel template defining all categories found in the database together with formatting guidelines. The latest version of the data extraction protocol can be downloaded here. Instructions for how to fill in the template are found in the template. The instructions are also available as a separate file that can be downloaded below.
The data extraction protocol contains around 400 different categories Those can be grouped into: reference data, cell related data, data for every functional layer in the device stack, i.e. substrate, electron transport layer, perovskite, hole transport layer, back contact, etc., synthesis related data for every layer, as well as key metrics related to IV, QE, stability, and outdoor measurements